
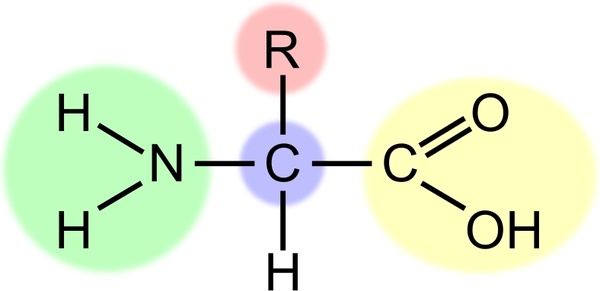
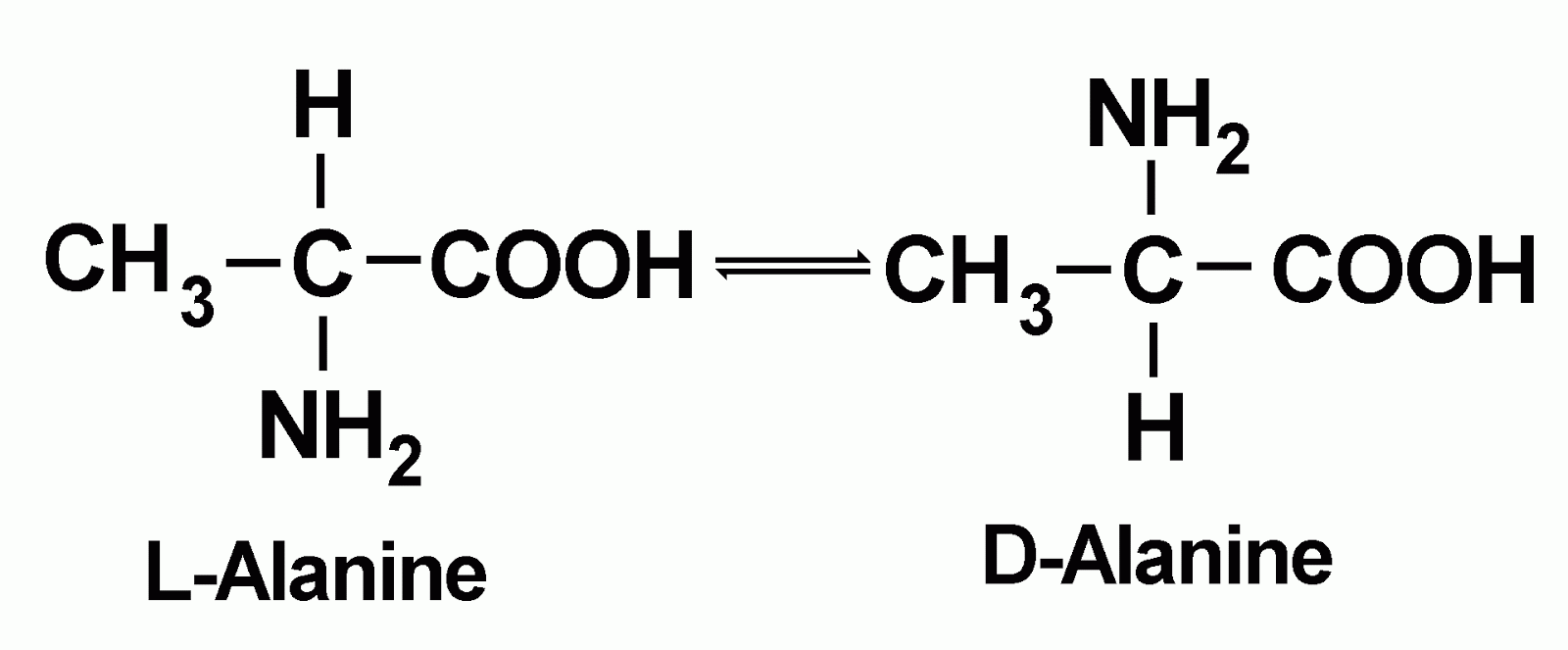
These cases, a "REMARK 465" entry will be included in the header of the PDBįile to identify each missing residue. Records, since the portion of the chain was present during the experiment. However, these amino acids will often be included in the SEQRES The ends of chains and mobile loops are often not observed inĬrystallographic experiments, and coordinates are not included as ATOM records In many cases, you may find that the coordinates presented inĪTOM records in a PDB file may not exactly match the sequence in the SEQRES SEQRES 4 A 63 GLU GLU LEU LEU SER LYS ASN TYR HIS LEU SEQRES 3 A 63 ARG LYS LEU GLN ARG MET LYS GLN LEU GLU SEQRES 2 A 63 ARG ALA ARG ASN THR GLU ALA ALA ARG ARG SEQRES 1 A 63 MET ILE VAL PRO GLU SER SER ASP PRO ALA
SEQRES 1 B 19 DT DG DG DA DG DA DT DG DA DC DG DT DC


 0 kommentar(er)
0 kommentar(er)
